Counting cells using image analysis with Marvin
Automatic cell counting methods is a powerful tool for biologists to understand how cells behave in some specific scenarios. Counting the number of cells from confocal microscopy samples might shows the bellance between cell division and cell death. This information is useful for studying how cells behave in many different scenarios including deseases and injury.
The example below demonstrates how to use image processing to count the number of cells from a given sample. Basically, the used approach works as follows:
- 1. Loads sample image containing many cells.
- 2. Increases image contrast between foreground and background elements.
- 3. Uses thresholding considering brightness neighborhood in order to separate foreground elements (cells) from the
background.
- 4. Separates grouped cells using morphological operations. The image is converted to the binary format before this process.
- 5. Segments each cell using flood fill segmentation approach. For each segment, get its blob, convert it to contour and outline
each cell.
- 6. Saves the output image.
- 7. Prints the number of cells.

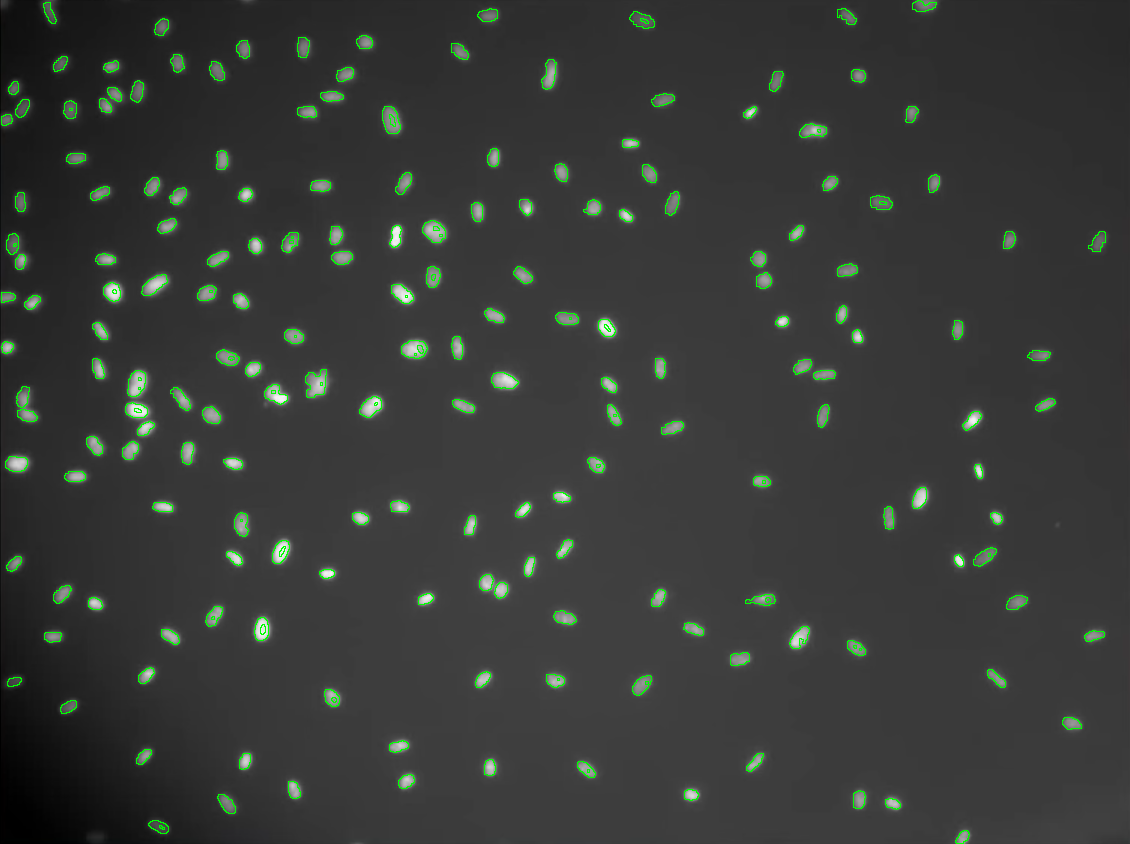
total cells: 184 Source:
package countCells; import static marvin.MarvinPluginCollection.*; import marvin.color.MarvinColorModelConverter; import marvin.image.MarvinBlob; import marvin.image.MarvinBlobSegment; import marvin.image.MarvinContour; import marvin.image.MarvinImage; import marvin.io.MarvinImageIO; import marvin.math.MarvinMath; import marvin.math.Point; public class CountingCells { public CountingCells(){ // 1. Load sample image MarvinImage originalImage = MarvinImageIO.loadImage("./res/cells.png"); MarvinImage image = originalImage.clone(); // 2. Increase contrast brightnessAndContrast(image, 0, 30); MarvinImage imageOut = image.clone(); // 3. Thesholding thresholdingNeighborhood(image, imageOut, 1, 8, 1); // 4. Separate cells that are grouped invertColors(imageOut); MarvinImage bin = MarvinColorModelConverter.rgbToBinary(imageOut, 50); morphologicalErosion(bin.clone(), bin, MarvinMath.getTrueMatrix(3, 3)); morphologicalDilation(bin.clone(), bin, MarvinMath.getTrueMatrix(3, 3)); // 5. Segment each cell image = MarvinColorModelConverter.binaryToRgb(bin); image.setAlphaByColor(0, 0xFFFFFFFF); MarvinBlobSegment[] segments = floodfillSegmentationBlob(image); int totalCells=0; for(MarvinBlobSegment s:segments){ MarvinBlob blob = s.getBlob(); MarvinContour contour = blob.toContour(); if(blob.getArea() > 50){ totalCells++; for(Point p:contour.getPoints()){ originalImage.setIntColor(s.getX()+p.x, s.getY()+p.y, 0xFF00FF00); } } } // 6. save output image MarvinImageIO.saveImage(originalImage, "./res/cells_output.png"); //7. Print the number of cells System.out.println("total cells: "+totalCells); } public static void main(String[] args) { new CountingCells(); } }